You may or may not know that this semester for the Choose Ohio First Bioinformatics scholarship I'm working with Andrew on RNA secondary structure prediction. If not, you do now ;) We've looked over some papers, and found a highly referenced work on the topic, a dynamic programming approach. I'm hoping to come back and make a post about the papers we read and specifically about the algorithm we're currently using. We were able to shortcut some steps because there is a provided reference implementation of the algorithm. We're still working on understanding it, but it should prove as a good starting point and potentially as something we might be able to extend. In addition to that, we've decided we would like to have a visualization of the folded RNA. Not just a static visualization, however, but one you can change to see how the overall energy changes
So, where do we start on something like that? Well, there is a pretty popular piece of software named jmol, which has visualisation. However, it has many more features than we need and because of that might be hard to extend. What we want is relatively simple, so right now we're both trying our hand at implementing it. Andrew is trying in java, and I'm doing it in C.
We don't get something that says "the 33rd guanine goes in these
coordinates", so we must come up with something ourselves. I thought about
it for a little bit, but couldn't come up with anything satisfactory. So,
instead I opted to start off with a Feynman diagram form. For this
visualization, the chain is laid out in a circle, and bonds between pairs
are arcs between them. This is really neat, because it is very easy to see
where there are various structures like hairpins and loops. The last few
hours was spent hacking away on it, and I've come up with something I think
looks pretty neat (and lets you modify the structure, the main purpose!).
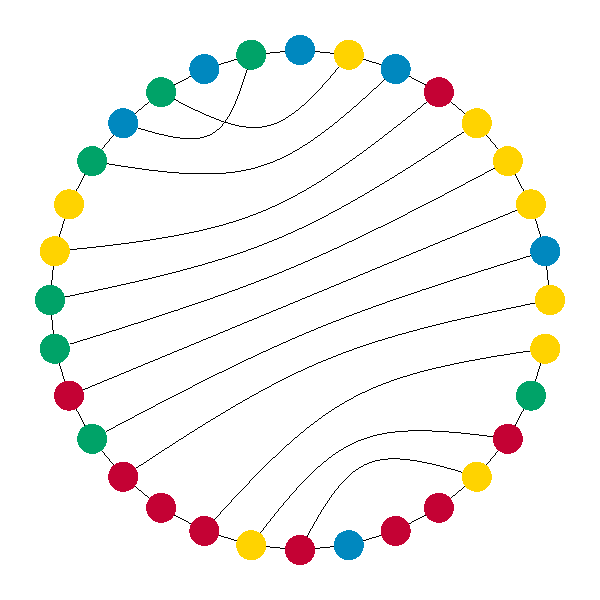
This is a slightly modified result of the optimal RNA secondary structure prediction algorithm, to show off more interesting structures. Before, there were some external bases and one loop, not interesting :) This is an actual screen-shot of the program, and the bonding modifications were made inside the program. The next thing I need to have is text rendering, to say what is selected, what is hovered over, and whether or not those can be paired. In addition, there will be a continuous energy rating somewhere. An idea I had is, once you select or hover over a base, background threads can start exploring modified structures based on altering that pair, and then give you "hints" if it finds a structure that is more optimal than what is there presently.
The main reason I wrote this post, though, is the weighted, arbitrary degree Bézier curve drawer I implemented for this. Basically, the arcs showing bonding between bases went through a few iterations. I knew what I wanted them to look like, but that was too complicated to start with. So, it started out as simple straight lines. The next thing I did was to figure out the "distance" around the circle the bases are (allowing jumping over the hole on the right). Then, I created a circle concentric to the backbone whose radius is inversely proportional to the distance between the bonded bases. Then, the point on that circle at the angle that was half way between the two bases was chosen as an intermediary point between the first and second base. Two straight line segments were drawn, connecting these three points. Finally, a weighted Bézier curve drawing function was implemented to interpolate the points with a weight of 1 for the endpoints and 2 for the midpoint. The Wikipedia article portion about the rational form was succinct, but easy to understand. To calculate the (n choose i)'s I simply started at 1 and calculated the next number to the right in Pascal's triangle by the obvious method (multiply by degree minus i, divide by i).